导图社区 DNA replication
- 36
- 1
- 0
- 举报
DNA replication
DNA replication:Initiation、Elongation、Terminations、some terms' defination、some enzymatic conventions……
编辑于2022-06-09 22:51:11- DNA repli…
- enzymatic
- Terminati…
- Elongation
- 相似推荐
- 大纲
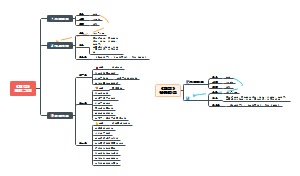
DNA replication
Initiation
initiation site
Prokaryotic
OriC
repeated nucleotides
bind initiator proteins
4*9-mer repeats
A-T rich sequence
3*13-mer repeats
open site
240bp/4Mb Ecoli genome
single replicon
initiator proteins
DnaA
DnaAs bind and wrap around 9-mer repeats make A-T rich sequence opens
Eukaryotic
no specific origins
may depends on both sequence and chromatin structure
0.5-55kbp intiation zone with no signal sequences
multiple replicons
no define number
devolopment dependent
increases with size of chromosome
initiator proteins
ORC(origin recognition complex)
six-subunit protein
Replisome
Helicase
role
Unwinds DNA double helix
produces torsional strain
cause supercoiling
Prokaryotic
DnaB
helicase loader(DnaC) loads 2 Helicase(DnaB) to the 2 replication folks of the opened A-T rich region
DNA replication is always bidirectional (exept some viruses)
loading requires 1 ATP per Helicase
forms prepriming complex
Structure
hexamer with 1.7nm hole
Eukaryotic
MCM
Mini Chromosome maintenance
Moves 5’ to 3’ on lagging strand template
with hydrolysis of ATPs
Topoisomerases
Type I
transiently nick one strand, pass the intact strand through the nick, then reseal the nicked strand.
Type II
break both strands and pass a double helical section through the double-strand break. They require ATP.
e.x.DNA gyrase
relieve supercoiling
DNA Primase
role
leading strand
synthesis once at initiation
lagging strand
synthesis periodically
Prokaryotic
only RNA polymerase
syntheses 10-20bp long RNA primers
Eukaryotic
Primase–Pol α complexes
contains both RNA & DNA polymerase
Primase synthesis first short RNA and extend with DNA by pol α
After each synthesis, Primase dissociates
DNA polymerase
Prokaryotic
DNA polymerase I
DNA synthesis
5'-to-3' DNA polymerase activity
proofreading
3'-to-5' exonuclease activity
larger fragment
res-324 to 928 = Klenow fragment
role
replaces primer with DNA
1. erases primer
5'-to-3' exonuclease activity
smaller fragment
res-1 to 323
2. fill gaps
DNA polymerase II
DNA synthesis
5'-to-3' DNA polymerase activity
proofreading
3'-to-5' exonuclease activity
role
DNA repair
DNA polymerase III
DNA synthesis
5'-to-3' DNA polymerase activity
proofreading
3'-to-5' exonuclease activity
role
main replication enzyme
subunitits
β subunit
provides high processivity
2 identical protein chains
form a circular sliding clamp
requires clamp lodder to open it, load it on DNA, and close it
DNA polymerase IV
DNA polymerase V
Eukaryotic
2 main replication enzymes
polymerase delta(pol δ)
PCNA-Rfc(Replication factor C)-Pol δ displaces primase-polα
increases processivity
increases efficiency
5 times faster than pol α
it's easier if you just view it as part of primer..
polymerase epsilon(pol ε)
Single strand binding proteins(SSB)
role
stabalize exposed hydrophilic base in aqueous environment
Eukaryotic
RPA proteins
sliding clamps
Prokaryotic
β subunit of DNA pol III
Ring shaped dimer
each monomer having three globular domains
Eukaryotic
PCNA (proliferating cell nuclear antigen)
discovery
because this protien exist in proliferating cell nucleus and induce antibody productin
structure
Ring shaped trimer
Clamp loading complex
role
loads clamps
1. loads a β subunit on DNA single strand
leading strand
load onece
lagging strand
load periodically
2. transfers DNA pol III to every loaded clamp
holds 2 DNA pol III together with other accessory proteins
structure
claw domain
catch and loads clamp
2 DNA pol III main body attaching domain
constantly attached to the main body
tail domain
binds to helicase on lagging strand
conserved arrangement in all 3 domains
clamp loader, sliding clamp, 2 polymerases, helicase, and primases at replication folk
Elongation
leading strand
single priming event
lagging strand
looping
make DNA pol IIIs on 2 strands(DNA pol dimmer) move in same direction, but DNA synthesis in two strands under different directions
because DNA polymerase III periodically dettaches
perioditity
Clamp
it detaches spontaneously when the elongating okazaki fragment reaches previous primer
Then this primer is removable by DNApol I
Clamp loader
it always holds 2 DNA pol III major subunit
it periodically catches a β clamp, encircle it on new primer site of DNA template and transfer the free DNA polIII body on it
DNA primase
periodically synthesis primers
DNA polymerase III
periodically dettaches from DNA template
because β clamp is periodically dettached
and main body dissociates with β clamp
and reassociates with β clamp next round
loop
grows
loop grows when DNApolIII elongating okazaki fragemts
refills
after the new dettach & loading event, the loop is refilled
Eukaryotic okazaki fragments maturation
No DNA polymease 1
so pol δ replaces the primer, then RPA covers the protruding and Dna2 enonuclease cleaves it, finally DNA ligase I stichs the nick using 1 molecule of ATP
Protein changes
primer removals
prokaryotic
DNA pol I
eukaryotic
pol δ displaces backward from 5' end
stich nicks/DNA ligase
prokaryotic
DNA ligase
1. NAD+
Eukaryotic
DNA ligase I
1 ATP
Terminations
termination site
Prokaryotic
Ter
conserved sequence: GTGTGTTGT
roughly opposite to oriC
Eukaryotic
telomere
elongated by telomerase
termination proteins
Prokaryotic
Tus
contra-helicase
DNA intertwining
only prokaryotic issue
opened by DNA gyrase
some terms' defination
Replicon
The DNA contolled by an origin
topological state of DNA
the coiling of double helix
torque
tortion strain
supercoiling
coil in space
intertwining
like two rings in chains
leading strand
continuous strand
lagging strand
discontinuous strand
Okazaku fragments
DNA fragments synthesized on the lagging strand
replication fork
the partial opening DNA helix, here replisome forms
processivity
The ability of a polymerase to remain attached to the template
Replisome
The assembly of enzymes involved in DNA replication
subdivision
primosome
primase complex with helicase
DNA pol III dimer
some enzymatic conventions
all polymerase synthesize nucleotides from 5' to 3' direction
all DNA polymerases require primers, but RNA polymerase don't