导图社区 Protein purification
- 17
- 0
- 0
- 举报
Protein purification
这是一篇关于Protein purification的思维导图,主要内容有The protein we are seperating、1.lon exchange chromatography、2.Gel filtration chromatography、3.Bradford protein concentration method等。
编辑于2022-06-11 15:55:32- Protein p…
- SDSpages
- Bradford
- 相似推荐
- 大纲
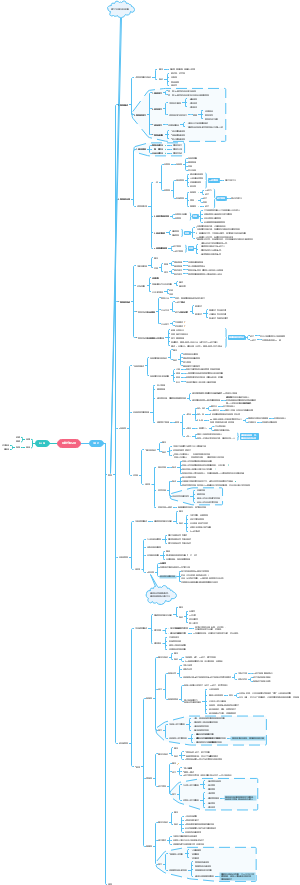
Protein purification
The protein we are seperating
cytochrome C, myoglobin and ferritin
all same reddish colour
Myoglobin
single polypeptide 16900
1. Ion exchange chromatography
principle: seperates proteins by charges
Colomn
Matrix
diethyl amino ethyl- (DEAE) on matrix is positively charged
so exchange anions
negatively charged would be drag down
100mM Tris-HCl: PH-9.0
Elusion
Elusion buffer: Nacl
best be increasingly high concentration
Salt gradience
Result
Flew through fraction
Cytochrome C
Elute fraction
Myoglobin
Ferritin
details
The column is equilibrated in 100 mM Tris-HCl, pH 9.0 and the pump is set at 1.0 mL min-1. Run the column as follows, but retain 50-100 μL of the original sample for analysis.
2. Gel filtration chromatography
Principle: seperate proteins by sizes
limitation: low resolusion due to diffusion
benifit: get nature protein
Pore size
Sephacryl, S200HR, will separate proteins in the range 5 - 250 kDa
Any protein larger than 250kDa will elute first (void volume)
So the big ferritin will first elute
Result
Peak 1
Ferritin
Peak 2
Myoglobin
3. Bradford protein concentration method
aim
assess the final purity and yield after seperation
procedure
identify 2 peaks first using the scanned absorbance
Scanning spectrophotometry
350nm-650nm
results in 426nm
measure the dye's absorbance at 595nm which accounts for the real protein concentration
Standard protein solusion
calibration curve
IgG solusion in 100mA Tris-HCl
280nm ab
The absorbance of a 1.0 mg mL-1 solution of IgG is 1.44 at 280 nm.
Bradford dye
1.0 mL of Bradford dye reagent is added to 20 μL of a protein sample, mixed and incubated at room temperature for at least 5 min
Color is from brown to red
ab of the dye is 595nm
actually the ab is scanned to find the peak in our proteins cases
blank produced by adding 1.0 mL of dye reagent to 20 μL of the appropriate buffer
Result
OR
0.55g/ml
FT
0.17g/ml
Elute
0.50g/ml
peak 1
0.085g/ml
Peak 2
0.046g/ml
4. SDSpages
Whole name
polyacrylamide gel electrophoresis
benifit: high resolusion(no diffusion)
limitation: dinatured and seperated subunits
polyarylamide gel
12% polyacrylamide
toxic
but it's more dense and able to seperate small molecular weight's proteins
Agrose gel can only seperates molecules with large MW
like DNA, RNA
otherwise for protein will diffuse
Principle: seperate proteins by MW
How to make the gel
Two kinds of gel with slightly different density therefore the proteins can condense at the interface
Both the gel, the loading dye buffer, and the buffer around the gel contains SDS
After mixed the sample with the loading die buffer
It heats in 100 degree for 3min to completely denature the protein.
And let the SDS attach around the linearized protein.
loaded samples
molecular weight marker
original mixture of the 3
IEC Elutted myoglobin+ferritin
IEC FT cytochrome c
GFC peak 1 ferritin
GFC peak 2 myoglobin
Commassie blue
To visualize the protein bands in our sample
5. Western blot
antibodies specific for each bar of SDS
Key words
Membrane
Proteins in SDS polyarylamide gel is transfered by layer of fliters and sponges under electric field
Subtopic
Blocker proteins
BSA or Milk
block the membrane part which not binds to the protein
reduce unspecific antibodies binding to membrane
Primary antibody
Recognize cytochrome c
Secondary antibody
recognize Fc region of the primary antibody
has HRP in its Fc region
an enzyme can oxidize a substrate and release light and the bands can be visualized under UV
Process of immune blocking
All these are done in a small box
first add blocker protein
wash
Don't wash too much, but gently wash out the blockers covered our target proteins
TTBS wash buffer
then add primary anitibody
Wash 3-5times, each time shake 5 min
wash to get rid of unspecific bindings
then add secondary antibody
Wash
6. Sequencing
+online database