导图社区 Method of Protein Regulations
- 12
- 0
- 0
- 举报
Method of Protein Regulations
这是一篇关于Method of Protein Regulations的思维导图,主要内容有Covalent changes、Reasons to regulate proteins、Noncovalent changes。
编辑于2022-06-11 16:05:46- Protein R…
- Protein
- Covalent …
- Noncoval…
- 相似推荐
- 大纲
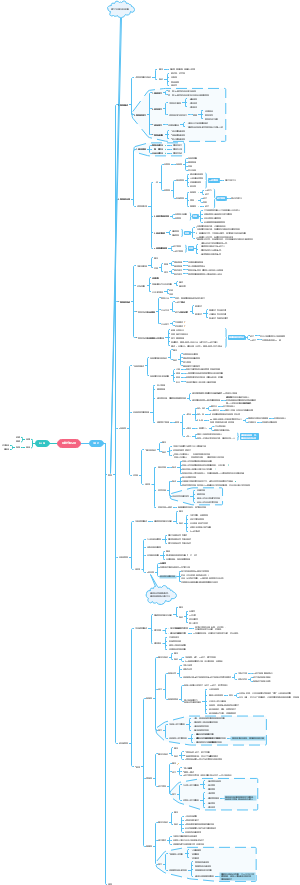
Method of Protein Regulations
Covalent changes
Cleavages/proteolysis
irreversible
by proteases
depend on cleavage partern
limited proteolysis
Site specific
activation
unlimited proteolysis
distructive
depend on residues of active site
Serine proteases
using a serine alcohol as nucleophilic attack
activated by histine
chymotrypsin
from chymotrypsinogen
synthesized in pancrease
first cleavage by trypsin
between arg15/leu16
into pi-chymotrypsin (active but not stable)
second cleavage
a-chemotrypsin (active and stable)
A,B,C 3 subunitss
Connected by
A-B disulfide bond
B-C disulfide bond
cleaves mainly on the carboxy side's amide bonds of
tyrosine
Y
tryptophan
W
phenylalanine
F
metheonine
M
rare
They contain aromatic ring/ big hydrophobic side chain
fit the enzymes' hydrophobic pocket
In enzyme "S1 binding pocket"
other aminoacids are cleaved in a slower rate
activity depends on actalytic triad including Serine 195
chemical modification reaction
28 possible ser
treat with DIPF
kinetics
measured by substrate analog with colored product
N-Acetyl-l-phenylalanine p-nitrophenyl ester
cleavage on ester bond
yield p-Nitrophenolate
yellow
2 phases
burst phase
steady state phase
catalysis mechanism
covalent
acylation & deacylation
Catalytic triad
Asp 102
stabalize His 57's left N-H
His 57
general acid/ base catalyst
Ser 195
nucleophilic acyl substitution of amide carbonyl
the carbonyl oxygen anion in terahedral intermediate is stabalized by the oxyanion hole
Serin's -OH as a nucleophile
alcohol lysis of amide
yield amine and ester
nitrogen of substrate's amide bond as a leaving group
regenerate base catalyst by protonate amine anion
ps. no intermediate, general catalyst
nucleophilic attack is generally catalyzed hy basic N of his57, by deprotonate the nucleophilic -OH
yield an acyl enzyme
serine is connected to the substrate
after that is the hydrolysis of the acyl enzyme(ester) by water
still general base catalyst by his57
deprotonate H2O to a better nucleophile
then hydrolysis of the ester
histidine protonates alkoxide ion as a better leaving group
regenerate base catalyst
ps. no intermediate, general catalyst
regenerate acid and alcohol
evolution
both homologous
chymorypsin family
and analogous
convergent evolution
ex. subtilisin
catalytic tiad
oxianion hole
Threonine proteases
using a threonine secondary alcohol
Cysteine proteases
using a cysteine thiol
Aspartate proteases
using an aspartate carboxylic acid
carboxylate act as general base catalyst and deprotonate water to initiate water's nucleophilic attack
Glutamic acid proteases
using a glutamate carboxylic acid
Metalloproteases
using a metal, usually zinc
metal cation stabalize water and let another base side chain to general deprotonate water to nucleophilic attack
depends on function
Nutrition lytic
act on most zymogens synthesized in pancrease
Invasion
Evasion
Adhesion
Processing
Signaling
Drug as protease inhibitor
carptopil
inhibit metalloprotease angiotensin-converting enzyme (ACE)
regulate blood pressure
Crixivan
inhibit HIV viral protease
contral HIV
mimix tetraheral intermediate
alcocol
other mimics S1,S2,S1',S2' binding sites
Self cleavage
self activation
NTN hydrolase family: lose internal peptides
ex.
activiation of zymogens
digestive enzymes
remove of propeptides
others enzymes removal of internal peptides
activation of hormones
tissue specific cleavage
a lot of hormones come from a single precorsor
blood clotting cascades
in signaling transduction pathway
caspases which activate apoptosis
procollagenase
collagenase
remodel collagen
in frog development
complex system:
proteolytic cascades involved
digestive cascades
blood clotting cascades
Substitutions
mostly reverisible
phosphorylation/dephosphorylation
by kinase
mosty activated with ATP binding to it
1.5-2.0% of the genome
& phoshpatase
ex.
metabolism
signaling transduction Kinase cascades
MAPK
mitogen activated protein kinase
MAPKKK>MAPKK>MAPK
signal amplification
transcribtion
RNApolII's movement
Act mostly on
Eukaryotic
Ser, Thr, Tyr
Prokaryotic
His, Asp
Actually most is kind of Allosteric
ex. kinase phosphorylate serine far from active site of glycogen phosphorylase, and inactivate it
acetylation
ex. histone
at lys or N-terminals
reversible
Methylation
On Arg and lys
epigenetics
irreversible?
Ubiquitination
target to proteosome degredation
lipid association
Myristoylation
amide bond links C14 lipid with protein N-terminus
Palmitoylation
thioester bonds link C16 lipid with cys residue
Reasons to regulate proteins
overcome low speed of translation
turn on/off can save energy of translation
contol activities in different compartments
Simply works
Noncovalent changes
All reversible
for molecules require to turn on/off repeatly
molecules bind to proteins
proteins
e.x. α 1-antitrypsin inhibit elastase
elastase break down elastin
together with collagen strenthen connective tissue
normally against pathogen
a lys of it binds almost irreverisbly to elastase active site(S1 pocket's Asp)
one kind of acute phase protein
normally stored in liver, effect in lungs
deficiency of the gene leads to pulmonary emphysema
smoking waken its effect by oxidize a methionine residue
emphysema
organic metabolites/signaling molecules
cAMP
cAMP act as a common secodary messenger
Receptors generate cAMP when hormones activate them
Adenyl cyclase
transmembrane protein
activated by GPCR's ATP binded Ga subunit
cAMP dependent protein kinase
Part of the regulatory subunit resembles a kinase substrate and occupies the catalytic site of catalytic subunit: like CRRC
by a pseudosubstrate sequence
cAMP binds to the regulatory subunit, weaken those interactions and release the catalytic subunit of kinases
like RR+2C
GTP binding proteins family
Ras
120 kDa
Timer
Upstream signaling protein SOS interacts with RasGDP and activates it by exchange its binding GDP into RasGTP
under the GTP binding conformation, N-terminal SH2-SH3-SH2 domains transmit signal towards down stream by interact with downstream proteins(effectors).
C-terminal GAP and other domains of Ras hydrolyisis Ras binding GTP to GDP, and inactive Ras protein from RasGTP to Ras GDP.
part of the mitogenic signal transduction pathway
Carcinogenesis
Oncogenes
regulated by Ras GTPase activating protein
Ran
ran help impoltin back to the cytoplasm
it's activated by one subunit of importin into GTP binding form
timer
Small GTPase subunit
20-40kDa
or GAP domains
GAP adds an extra residue to the Ras GTPase catalytic site
accelerate timmer
metabolites
Usually feedback inhibition
simple competative
Allosteric
ex. Aspartate transcarbamoylase
inhibited by its product CTP
small ions
Ca2+
calmodulin
Ca2+ can activate it by conformational changes of allosteric binding , and activated calmodulin can further activate other proteins, like myosin kinase
myosin kinase phosphorylates myosin light chain, which leads to musle contraction.
Ca2+ act as another common secondary messenger
Fe2+
Diphtheria toxin gene repressor
Fe2+ binding moves DNA recognition helix close together, and moves away N-terminal blockage of DNA binding
proton
PH dependent conformational changes
usually utilized by parasites(fusion protein)
Dipthria toxin
reducing PH in endsome fasilitate releasing of toxic enzymatic domain
modify ribosome
low PH changes the conformation of the fusion protein: the translocation domain
fuse with membrane and release toxic domain
degestive enzyme
acid activates pepsin
protonates active site's aspartic acid
then self cleavage
double insurance
Allosteric
binding distance from active site
lead to conformational change
Competative
direct blockage of active site
combining
Glycogen phosphorylase
active enzyme/ inactive enzyme is proportional to [AMP]/[ATP}
regulated by allosteric activation, inhibition.
and phosphylation
Changes by regulation
change proteins' interactions with other molecules
substrate
enzyme
by
1. reorganize existing active site residues
most
by allosteric conformational changes
by cleavage
e.x. chemotrypsin
2. remove/add blockage on active site
3. change on active site
Formation
NTN hydrolase self cleavage's N-terminal
Blockage
Kinase phosphorylate a serine residue around the active site of isocitrate dehydrogenase
Phosphate group repels negative charged substrate
active site can be
catalytic site for enzymes
interaction sites for other proteins
DNA
DNA binding proteins
other proteins